Help:Guidelines EditorPalette
From WikiPathways
(→Cellular compartments) |
(→Data nodes) |
||
| Line 76: | Line 76: | ||
* <b>Pathway</b>: Should be used instead of a label to denote a connection to another pathway. | * <b>Pathway</b>: Should be used instead of a label to denote a connection to another pathway. | ||
[[Image:DataNodes.png|border]] | [[Image:DataNodes.png|border]] | ||
| + | |||
| + | [[Image:DataNode-RNA.png|border]] | ||
= Graphical elements = | = Graphical elements = | ||
Revision as of 19:32, 15 April 2014
This guide describes the most common uses of various elements of the WikiPathways editor palette. Examples are taken directly from the WikiPathways archives. If you have questions about specific cases, contact the discussion mailing list.
Interactions
Line / Arrow
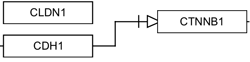
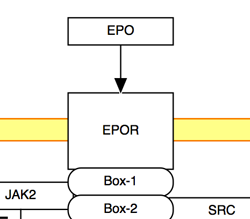
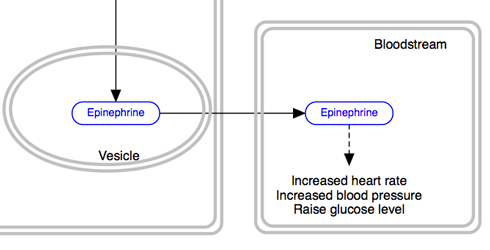
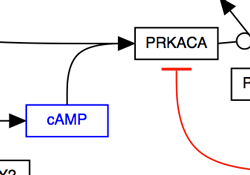
The solid line and arrow are used to denote a variety of processes, including conversion, translocation, activation, binding and modification.
| Enzymatic conversion | Receptor binding | Translocation between compartments | Activation by cAMP |
|---|---|---|---|
| 
| 
| 
|
Dashed line / dashed arrow
The dashed line and arrow are used to denote an uncertain process or a process that involves additional steps not outlined in the current diagram.
| Multi-step process without details |
|---|

|
T-bar
The T-bar is used to denote inhibition.
Line types
Any interaction can have a line style of either straight, curved, elbow or segmented. The default style is straight, and is shown in the examples above. The other three line styles are used primarily to increase readability of complex pathways.
| Curved | Elbow | Segmented |
|---|---|---|
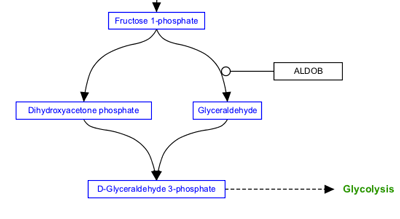
| 
| 
|
Interactions - Molecular Interaction Maps (MIM)
The WikiPathways editor includes a set of MIM interaction types, based on the Molecular Interaction Maps notation.
Necessary stimulation
Binding
Conversion
Stimulation
Catalysis
Transcription-Translation
Data nodes
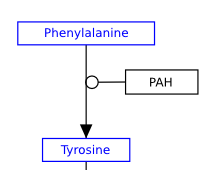
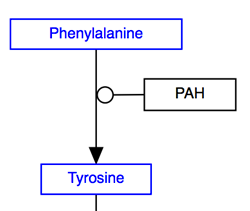
The below image shows the preferred usage for the most common data node types:
- GeneProduct: Default data node and should be used for any gene product.
- Metabolite: Should be used for any metabolite, drug or small molecule.
- Pathway: Should be used instead of a label to denote a connection to another pathway.