Portal:Semantic Web
From WikiPathways
| Line 23: | Line 23: | ||
{{{{FULLPAGENAME}}/box-normal | {{{{FULLPAGENAME}}/box-normal | ||
|<!-- TITLE OF BOX --> | |<!-- TITLE OF BOX --> | ||
| - | <big> | + | <big>Open PHACTS API</big> |
|<!-- EDIT LINK --> | |<!-- EDIT LINK --> | ||
| - | {{FULLPAGENAME}}/ | + | {{FULLPAGENAME}}/Open_PHACTS_API |
|<!-- CONTENT LINK --> | |<!-- CONTENT LINK --> | ||
| - | {{FULLPAGENAME}}/ | + | {{FULLPAGENAME}}/Open_PHACTS_API |
|<!-- FOOTER LINK --> | |<!-- FOOTER LINK --> | ||
}} | }} | ||
Revision as of 08:05, 5 May 2017
The WikiPathways Semantic Web Portal
Welcome to the WikiPathways Semantic Web Portal!
This portal describes the Semantic Web features of the WikiPathways databases, such as the Resource Description Framework (RDF) translation, the ontology, and the new nanopublications.
The WikiPathways RDF is provided as part of the monthly releases and contains the Curated and Reactome pathways. The RDF is split in two parts, the GPMLRDF part which contains a direct translation of the content in the GPML files, and a WPRDF part which contains the biology represented in the GPML
The WikiPathways Vocabularies
The WikiPathways vocabularies are for the semantic information about the pathway, data nodes, and interactions and the GPML vocabulary is for the graphical information about how the pathway diagram is laid out and represented.
How to cite
If you use the RDF, vocabularies, or nanopublication, please cite the following paper:
- Waagmeester, A., Kutmon, M., Riutta, A., Miller, R., Willighagen, E. L., Evelo, C. T., Pico, A. R., Jun. 2016. Using the semantic web for rapid integration of WikiPathways with other biological online data resources. PLoS Comput Biol 12 (6), e1004989+. doi:10.1371/journal.pcbi.1004989 . For the pathway content, please follow these How to cite WikiPathways instructions.
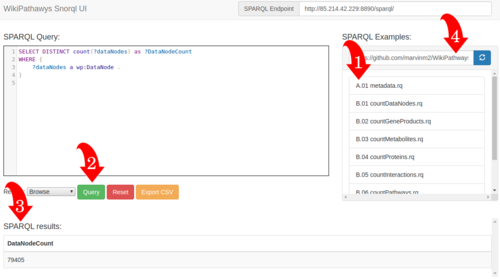
Snorql Interface
Visit our new Snorql interface at sparql.wikipathways.org. The image to the left explains which steps you can take: 1: Select a query from the list. 2: Press the green query button to execute your selected query. 3: View the results on the same page. 4: You can select your own list of example queries from github, by adding the link.
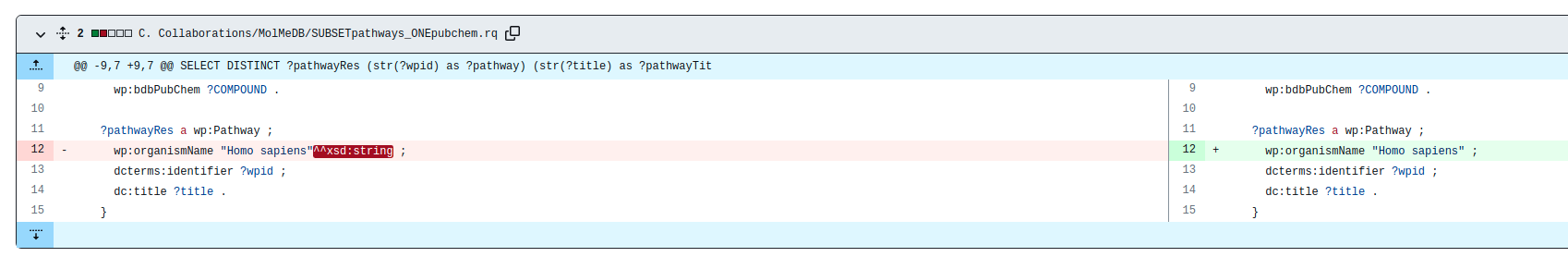
NOTIFICATION: Due to an Apache update, we are now creating RDF data according to SPARQL 1.1. However, our SPARQL-endpoint running on Virtuoso is still using SPARQL 1.0. This influences the way to query strings, and might affect federated queries. Please remove the ^^xsd:string suffix, as shown in the example below.
News
2021-02-16 - Our Snorql interface now has a feature to get a permanent link to a query.
2020-08-12 - Our endpoint now has a NEW Snorql interface!
2020-06-01 - Because OpenRiskNet cloud was not back yet, an alternative location was found.
2020-05-14 - Since this Wednesday the OpenRiskNet cloud where the SPARQL endpoint has been hosted is showing network issues. It is being explored.
2020-03-31 - The first COVID-19 pathways are now available in the RDF. We will try to update the RDF weekly for these.
2020-01-08 - A solution has been put in place, thanks to the OpenRiskNet project. The current SPARQL endpoint resides at http://sparql.wikipathways.org/sparql
2019-12-24 - Since the University of Maastricht is suffering from a cyber attack, our endpoint is not reachable. We're currently working on a solution!
2019-12-17 - The enpdoint is updated with December data, including additional quality checks with our Unit Tests.
2019-11-21 - The SPARQL enpdoint is updated with the November data.
2019-10-10 - The SPARQL enpdoint is updated every month with the Approved and Reactome data. We are supporting metabolite mappings to ChEBI, Chemspider, HMDB, LipidMaps, KEGG Compound, PubChem and Wikidata!
2018-11-18 - Five species have been added to the RDF: Equus caballus, Mycobacterium tuberculosis, Plasmodium falciparum, Populus trichocarpa, and Solanum lycopersicum.
2018-10-12 - Paper about how WikiPathways RDF is used to expose interactions via Open PHACTS
2018-01-01 - The RDF is being used to expose facts from WikiPathways as nanopublications.
2017-05-04 - This portal was created
2016-06-23 - Andra Waagmeester's Using the Semantic Web for Rapid Integration of WikiPathways with Other Biological Online Data Resources paper is published
Open PHACTS API
Human approved pathway data is now also part of Wikidata! Check out which federated queries you can perform.
Downloads
The Semantic Web WikiPathways comes in two flavors: as RDF (beta) and as nanopublications (very experimental).
The RDF
You can download the WikiPathways RDF from here.
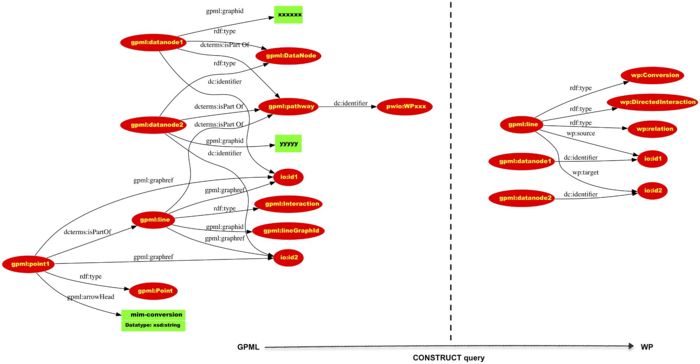
The WikiPathways RDF is split in two parts, the GPMLRDF part which contains a direct translation of the content in the GPML files, and a WPRDF part which contains harmonized biological information present in the GPML.
This figure from the 2016 paper shows how the WPRDF can be derived from the GPMLRDF:
SPARQL
We provide a SPARQL endpoint where data queries can be done.
WikiPathways Example SPARQL Queries
We have a large collection of general example queries and metabolite related example queries.
For example, to list all pathways per instance of a particular gene or protein (wp:GeneProduct), you can use the following SPARQL:
PREFIX wp: <http://vocabularies.wikipathways.org/wp#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX dcterms: <http://purl.org/dc/terms/>
SELECT DISTINCT
?pathway
(str(?label) as ?geneProduct)
WHERE {
?geneProduct a wp:GeneProduct .
?geneProduct rdfs:label ?label .
?geneProduct dcterms:isPartOf ?pathway .
?pathway a wp:Pathway .
FILTER regex(str(?label), "CYP").
}
Support
The research leading to these results has received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement no. 115191, resources of which are composed of financial contribution from the European Union's Seventh Framework Programme (FP7/2007-2013) and EFPIA companies’ in-kind contribution.