Help talk:Guidelines
From WikiPathways
Contents |
Graphical style guideline discussions
Multiple similar interactions
How should we display multiple similar interactions in such a way that it looks nice while the interactions are correctly represented? An example is in the Smooth Muscle Contraction pathway:

You see that the interactions to the GNAS protein are grouped by an arc. However, if we just link the lines to the arc, like this:

Then the interactions will not be recognized properly (since there is an interaction with the arc, not with GNAS).
Alternatively, we could explicitely draw each interaction seperately, but as you can see below, this doesn't look nice:

Any ideas on how to solve this?
- Thomas 17:54, 14 May 2008 (UTC)
Can we have each of the separate lines from the receptors link to a connector hub? I'm not sure if this edge connector type exists, but I've seen it illustrated at:
http://www.wikipathways.org/index.php/Pathway:Rattus_norvegicus:Catecholamine_synthesis
This would seem to solve the problem if you can have multiple lines merge into a single line and have anchor points on that line, so that an inhibition edge hits that. I was able to make this in WP, but just to an anchor so it's not a valid hub. I can take or leave the brackets if something like this works.

- Nsalomonis 23:13, 18 May 2008 (PST)
Looks good to me. I went ahead and made the change to the human version of the pathway here so folks can check it out: http://www.wikipathways.org/index.php/Pathway:Homo_sapiens:Smooth_muscle_contraction
-AlexanderPico 07:18, 19 May 2008 (UTC)
Looks great! I'd say this is the perfect solution as visual representation.
The only thing I'm still concerned about is that the line that points to Gnas has a start point that doesn't connect to anything. So what does it mean? (think about trying to convert it to Cytoscape).
I think this demonstrates a flaw in the underlying data model of line anchors. There are now three uses for line anchors, so their meaning is ambiguous:
- A tool to allow representation of mediators of an interaction
- A tool to allow multiple input/output for a reaction
- A tool to group similar interactions (connector hub)
The line anchors are designed to work well for the first use case. But the model isn't optimal for the latter use cases. For example, if you look more closely at the Catecholamine synthesis pathway, you'll see that the way metabolic reactions are stored is a bit odd. There is a single line which start point is connected to one of the substrate and which end point is connected to one of the products. But which substrate to choose if there are multiple? The other substrates are connected to the line that represents the reaction, but do they have a different meaning than the substrate to which the line is directly connected? And how to make a distinction between the enzyme and the reaction input/output? I'm not sure if in the end, we have enough information to derive the real meaning from the current representation. For now, we should just continue to use anchors for all these purposes and use Nathans representation for the problem of similar interactions, because it's certainly improving the pathways. But we should really start defining the actually meaning and find out how to represent it better. One solution we already planned is to allow the user to define interaction type of an edge, but I see now that it doesn't solve everything.
- Thomas 09:30, 19 May 2008 (UTC)
Right. It is fair to say that we are not ready to represent each distinct type of relationship in full detail. We will want to follow the BioPAX roadmap in developing our semantics and then fine tune our visual representations (including anchors) to serve the semantics and human readability. In the meantime, it's worthwhile connecting up pathways with the "smart connectors" we have now, even if imperfect, making the work easier later, perhaps something we can even script.
Here's how I would represent Catecholamine synthesis to clarify the flow of the biochemistry:
http://www.wikipathways.org/index.php/Pathway:Rattus_norvegicus:Catecholamine_synthesis
-AlexanderPico 03:30, 20 May 2008 (UTC)
How should we represent stoichiometry in pathways
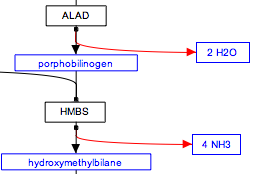
Should stoichiometry of reactions be displayed in pathways? At least one pathway (Heme Biosynthesis) has this information encoded in labels. While editing, I switched some of these labels to metabolites, and in doing so had the following choices:
- Represent the stoichiometry by changing the text label of the metabolite,for example from "water" to "2 H2O".
- Disregard the previous information and simply keep the text label without the stoichiometry, for example "H2O.
See the following figures for the difference. Option 1:
Option 2:
The first of these options didn't seem correct to me, so I ended up with option 2. We could also consider a third option of encoding this information in the comments of the edge (or node?).
Comments?
- Khanspers 23:11, 23 May 2008 (UTC)
Actually, I'd prefer option 1 until we support storing this information elsewhere. In option 1 the stoichiometry information is completely lost, while in option 2 it's still available as human readable information. And since the text label of a metabolite box is independent of its annotation (if you change 'H2O' to '2 H2O', it still points to H2O in the database), there is no 'computable' information lost. What do you think?
- Thomas 13:21, 28 May 2008 (UTC)
There is one more option, representing each molecule separately:
This could get messy, but I thought it should be represented here for completeness. Even though option 1 doesn't seem entirely correct (change the label on a metabolite from H20 to 2H2O), I think Thomas has a point. At least it is human readable and doesn't interfere with the database link. So for now I will change the Heme Biosynthesis pathway to reflect this.
How should we use ligand/receptor pairs?
The ligand and receptor line ends make for nice illustrations, but it is not obvious how to use these for linking objects. See the following example, from the inflammatory response pathway:
In this pathway, the ligand and receptor line ends are used mostly for decoration, for example receptor line for Il4ra is used to signify that this is a receptor. However, the receptor end of the line doesn't actually connect to anything. Also, placing the gene product over the line could erroneously be interpreted as the entity catalyzing a reaction. While editing this pathway, I have changed the representation into the following:
Do we have any good examples of how ligand/receptor pairs (or either alone) can be used in conjunction with connectors?
- Khanspers 23:11, 23 May 2008 (UTC)
A possible solution could be to make ligand/receptor pairs single line ends. So the ligand and receptor will be connected by a single connector, but the line end will be drawn as a ligand/receptor pair to make it look like the first image. Would that be a good idea?
Thomas 13:36, 28 May 2008 (UTC)
That's a good idea. This goes along with the general re-design of line ends proposed in an email.