Portal:Semantic Web/Intro
From WikiPathways
Current revision (15:29, 8 April 2022) (view source) |
|||
| (7 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | == Welcome to the WikiPathways Semantic Web Portal! == | |
| + | |||
| + | [[Image:newSnorqlInterface.png|500px|thumb|left|NEW Snorql Interface for SPARQL Endpoint]] | ||
[[Image:Sebweblogo.jpg|right|x150px]] | [[Image:Sebweblogo.jpg|right|x150px]] | ||
| - | This portal describes the Semantic Web features of the WikiPathways databases, such as the Resource Description Framework translation, the ontology, and the new nanopublications. | + | This portal describes the Semantic Web features of the WikiPathways databases, such as the [http://www.w3.org/RDF/ Resource Description Framework] (RDF) translation, the ontology, and the new nanopublications. |
| + | |||
| + | The WikiPathways RDF is provided as part of the monthly releases and contains the [http://wikipathways.org/index.php?title=Special%3ABrowsePathways&browse=---&tag=Curation%3AAnalysisCollection&view=thumbs Curated] and [http://wikipathways.org/index.php?title=Special%3ABrowsePathways&browse=---&tag=Curation%3AReactome_Approved&view=thumbs Reactome] pathways. The RDF is split in two parts, the GPMLRDF part which contains a direct translation of the content in the GPML files, and a WPRDF part which contains the biology represented in the GPML | ||
| + | |||
| + | === The WikiPathways Vocabularies === | ||
| + | |||
| + | The [http://vocabularies.wikipathways.org WikiPathways vocabularies] are for the semantic information about the pathway, data nodes, and interactions and the GPML vocabulary is for the graphical information about how the pathway diagram is laid out and represented. | ||
| + | |||
| + | == How to cite == | ||
| + | |||
| + | If you use the RDF, vocabularies, or nanopublication, please cite the following paper: | ||
| + | |||
| + | * Waagmeester, A., Kutmon, M., Riutta, A., Miller, R., Willighagen, E. L., Evelo, C. T., Pico, A. R., Jun. 2016. Using the semantic web for rapid integration of WikiPathways with other biological online data resources. PLoS Comput Biol 12 (6), e1004989+. doi:[https://doi.org/10.1371/journal.pcbi.1004989 10.1371/journal.pcbi.1004989] . For the pathway content, please follow these [[How to cite WikiPathways]] instructions. | ||
| + | |||
| + | == Snorql Interface == | ||
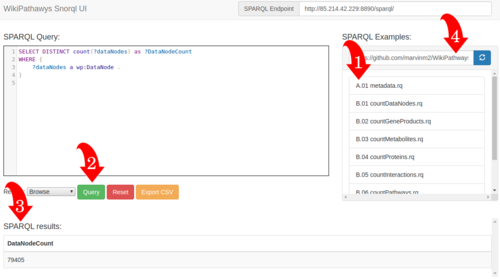
| + | Visit our new Snorql interface at [http://sparql.wikipathways.org/ sparql.wikipathways.org]. The image to the left explains which steps you can take: 1: Select a query from the list. 2: Press the green query button to execute your selected query. 3: View the results on the same page. 4: You can select your own list of example queries from github, by adding the link. | ||
| + | |||
| + | |||
| + | <pre style="color: red"> | ||
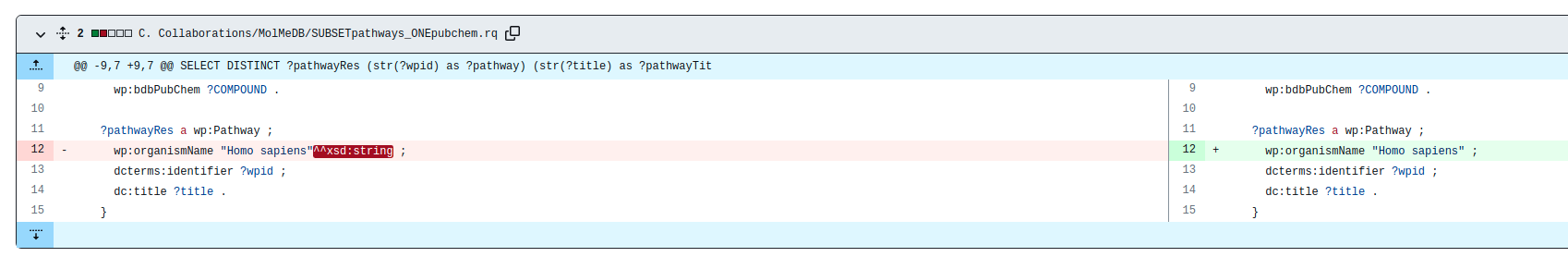
| + | NOTIFICATION: Due to an Apache update, we are now creating RDF data according to SPARQL 1.1. | ||
| + | |||
| + | However, our SPARQL-endpoint running on Virtuoso is still using SPARQL 1.0. | ||
| + | |||
| + | This influences the way to query strings, and might affect federated queries. | ||
| - | + | Please remove the ^^xsd:string suffix, as shown in the example below. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | </pre> | |
| - | + | [[Image:SPARQL11.png]] | |
Current revision
Contents |
Welcome to the WikiPathways Semantic Web Portal!
This portal describes the Semantic Web features of the WikiPathways databases, such as the Resource Description Framework (RDF) translation, the ontology, and the new nanopublications.
The WikiPathways RDF is provided as part of the monthly releases and contains the Curated and Reactome pathways. The RDF is split in two parts, the GPMLRDF part which contains a direct translation of the content in the GPML files, and a WPRDF part which contains the biology represented in the GPML
The WikiPathways Vocabularies
The WikiPathways vocabularies are for the semantic information about the pathway, data nodes, and interactions and the GPML vocabulary is for the graphical information about how the pathway diagram is laid out and represented.
How to cite
If you use the RDF, vocabularies, or nanopublication, please cite the following paper:
- Waagmeester, A., Kutmon, M., Riutta, A., Miller, R., Willighagen, E. L., Evelo, C. T., Pico, A. R., Jun. 2016. Using the semantic web for rapid integration of WikiPathways with other biological online data resources. PLoS Comput Biol 12 (6), e1004989+. doi:10.1371/journal.pcbi.1004989 . For the pathway content, please follow these How to cite WikiPathways instructions.
Snorql Interface
Visit our new Snorql interface at sparql.wikipathways.org. The image to the left explains which steps you can take: 1: Select a query from the list. 2: Press the green query button to execute your selected query. 3: View the results on the same page. 4: You can select your own list of example queries from github, by adding the link.
NOTIFICATION: Due to an Apache update, we are now creating RDF data according to SPARQL 1.1. However, our SPARQL-endpoint running on Virtuoso is still using SPARQL 1.0. This influences the way to query strings, and might affect federated queries. Please remove the ^^xsd:string suffix, as shown in the example below.