Help:Viewing Pathways
From WikiPathways
Current revision (19:10, 30 August 2021) (view source) m (Protected "Help:Viewing Pathways" [edit=sysop:move=sysop]) |
|||
| (45 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | __NOTOC__ | |
| - | __NOTOC__ | + | |
| - | + | Each pathway at WikiPathways has its own dedicated '''Pathway Page'''. In addition to the graphical representation of the pathway, they pathway page also includes other information about the pathway. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | == Graphical representation == | |
| - | + | === Interactive pathway viewer === | |
| + | The graphical representation of the pathway is located at the top of the '''Pathway Page''', as an interactive viewer: | ||
| + | * You can drag the pathway to pan and scroll to zoom. To reset pan or zoom, click the '''Reset''' button in the lower right corner. | ||
| + | * Click on any pathway element to display its annotations in a pop-up annotations panel. | ||
| + | * The annotations panel is organized as a list of data sources and contains link-outs to all cross-references for the xref. | ||
| + | * To find other pathways with the selected element, click the '''Find other pathways containing...''' link at the bottom of the annotation panel. This initializes a search at WikiPathways for the particular element. | ||
| + | * Move the annotations panel by click-and-drag on the '''move''' icon in the upper left corner, and use the '''close''' icon in the upper right corner to close the panel. | ||
| - | + | === Data formats === | |
| - | + | The pathway data can be downloaded in several formats using the '''Download''' button under the pathway image: | |
| + | * PNG, PDF and SVG image formats | ||
| + | * Gene list (.txt) format | ||
| + | * GPML format (GenMAPP Pathway Markup Language), an XML for annotated, curated pathway content. GPML-formatted files can be used in [http://www.pathvisio.org PathVisio] and [http://www.cytoscape.org Cytoscape]. | ||
| + | * PWF format, for use in the [http://www.ducciocavalieri.org/dct.htm| Eu.Gene ] application. | ||
| + | Batch download of pathways in gpml, svg, and rdf formats is available here: [[Download_Pathways |Batch Download]]. Additional formats are available as [[Daily_Download|Daily Downloads]]. A flat file of all pathways is also available on the batch download page in both html and txt format. | ||
| + | * BioPAX level 3 format (OWL) | ||
| - | + | == Quality Tags == | |
| - | + | '''Quality Tags''' are a set of tags that can be added to a pathway to notify the author and community about the state of the pathway. For example, you could tag a pathway that isn't annotated properly or tag your own pathway as work in progress. For more information on Quality tags and how to add them, please see the [[:Help:QualityTags|Quality Tags]] page. | |
| - | + | == Description == | |
| - | + | The '''Description''' section contains a description the pathway, entered by the author. The description can be edited by clicking the '''Edit''' link on the right side of the '''Description''' section. | |
| - | + | ||
| - | + | == Ontology Tags == | |
| - | + | The '''Ontology Tags''' section contains any ontology tags added to the pathway, and an interface for adding ontology tags. Ontology tags can be located through a search interface and browsable trees for the supported ontologies. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | == Bibliography == | |
| - | + | The '''Bibliography''' section contains relevant references for the pathway, from literature, textbooks, the web and other sources. The bibliography can be edited in the pathway editor, PathVisio. In PathVisio, right-click anywhere on the pathway drawing board, or on a pathway element, and choose '''Literature -> Add literature reference'''. | |
| - | </ | + | == History == |
| + | The '''History''' section provides detailed information about the changes made to the pathway. The pathway history is formatted as a table, with information on when the change was made, who made the change, and any comments entered by the user. A direct link to the pathway change is available by clicking the '''view''' link for each change. A '''revert''' link is also available for reverting the pathway to a previous version.<br> | ||
| + | |||
| + | == External references == | ||
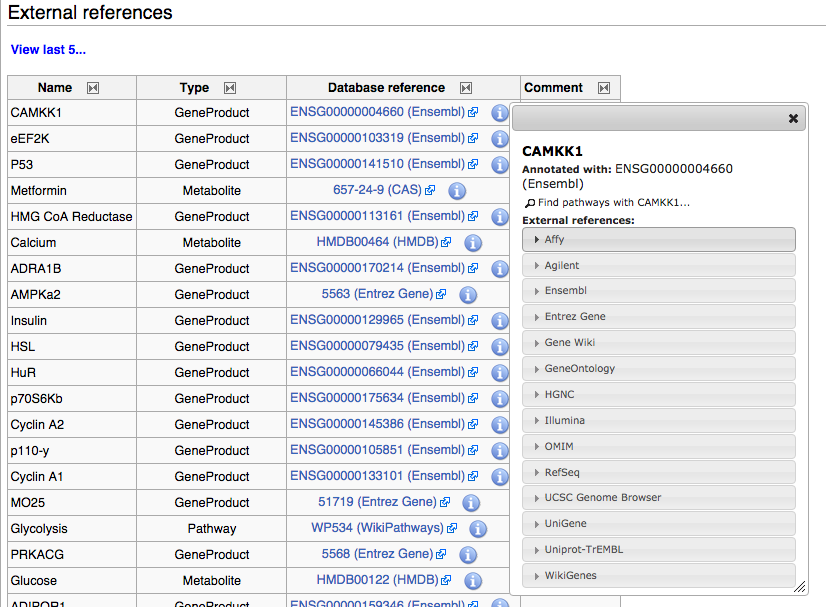
| + | The '''External references''' section contains lists of annotated genes, proteins, metabolite and pathway nodes, and interactions, that are on the pathway. Annotation on nodes and interactions are displayed in separate tables, titled '''DataNodes''' and '''Annotated Interactions'''. You can click the info icon (see screenshot below) to show a dialog containing links to many different resources that provide detailed information. | ||
| + | |||
| + | {{Help:XrefPanel}} | ||
| + | |||
| + | == Highlighting data nodes on pathways == | ||
| + | |||
| + | You can specify elements that you want to highlight on a pathway using a url with the following syntax: | ||
| + | |||
| + | http://www.wikipathways.org/wpi/PathwayWidget.php?id=WP528&label=PEMT&colors=green | ||
| + | |||
| + | For more information on highlighting of specific nodes on pathways, see the [[PathwayWidget|PathwayWidget]]. | ||
| + | |||
| + | <p>[[:Help:Contents|Return to Help Contents]]</p> | ||
| + | <p>[[:Help:Editing Pathways|Continue to Editing Pathways]]</p> | ||
Current revision
Each pathway at WikiPathways has its own dedicated Pathway Page. In addition to the graphical representation of the pathway, they pathway page also includes other information about the pathway.
Graphical representation
Interactive pathway viewer
The graphical representation of the pathway is located at the top of the Pathway Page, as an interactive viewer:
- You can drag the pathway to pan and scroll to zoom. To reset pan or zoom, click the Reset button in the lower right corner.
- Click on any pathway element to display its annotations in a pop-up annotations panel.
- The annotations panel is organized as a list of data sources and contains link-outs to all cross-references for the xref.
- To find other pathways with the selected element, click the Find other pathways containing... link at the bottom of the annotation panel. This initializes a search at WikiPathways for the particular element.
- Move the annotations panel by click-and-drag on the move icon in the upper left corner, and use the close icon in the upper right corner to close the panel.
Data formats
The pathway data can be downloaded in several formats using the Download button under the pathway image:
- PNG, PDF and SVG image formats
- Gene list (.txt) format
- GPML format (GenMAPP Pathway Markup Language), an XML for annotated, curated pathway content. GPML-formatted files can be used in PathVisio and Cytoscape.
- PWF format, for use in the Eu.Gene application.
Batch download of pathways in gpml, svg, and rdf formats is available here: Batch Download. Additional formats are available as Daily Downloads. A flat file of all pathways is also available on the batch download page in both html and txt format.
- BioPAX level 3 format (OWL)
Quality Tags
Quality Tags are a set of tags that can be added to a pathway to notify the author and community about the state of the pathway. For example, you could tag a pathway that isn't annotated properly or tag your own pathway as work in progress. For more information on Quality tags and how to add them, please see the Quality Tags page.
Description
The Description section contains a description the pathway, entered by the author. The description can be edited by clicking the Edit link on the right side of the Description section.
Ontology Tags
The Ontology Tags section contains any ontology tags added to the pathway, and an interface for adding ontology tags. Ontology tags can be located through a search interface and browsable trees for the supported ontologies.
Bibliography
The Bibliography section contains relevant references for the pathway, from literature, textbooks, the web and other sources. The bibliography can be edited in the pathway editor, PathVisio. In PathVisio, right-click anywhere on the pathway drawing board, or on a pathway element, and choose Literature -> Add literature reference.
History
The History section provides detailed information about the changes made to the pathway. The pathway history is formatted as a table, with information on when the change was made, who made the change, and any comments entered by the user. A direct link to the pathway change is available by clicking the view link for each change. A revert link is also available for reverting the pathway to a previous version.
External references
The External references section contains lists of annotated genes, proteins, metabolite and pathway nodes, and interactions, that are on the pathway. Annotation on nodes and interactions are displayed in separate tables, titled DataNodes and Annotated Interactions. You can click the info icon (see screenshot below) to show a dialog containing links to many different resources that provide detailed information.
Highlighting data nodes on pathways
You can specify elements that you want to highlight on a pathway using a url with the following syntax:
http://www.wikipathways.org/wpi/PathwayWidget.php?id=WP528&label=PEMT&colors=green
For more information on highlighting of specific nodes on pathways, see the PathwayWidget.