Portal:IEM/PathwayAnalysis
From WikiPathways
Current revision (12:53, 21 April 2022) (view source) |
|||
| (One intermediate revision not shown.) | |||
| Line 1: | Line 1: | ||
[[Image:NetworkAnalysisPurine.png|left|350px]] | [[Image:NetworkAnalysisPurine.png|left|350px]] | ||
| - | With the pathways from the Blau book (as well as with other pathways on WikiPathways), various data analysis approaches can be performed. | + | With the pathways from the Blau book (as well as with other pathways on WikiPathways), various data analysis approaches can be performed. You can find the relevant book chapter as Open Access regarding these data approaches here: Slenter, D.N., Kutmon, M., Willighagen, E.L. (2022). WikiPathways: Integrating Pathway Knowledge with Clinical Data. In: Blau, N., Dionisi Vici, C., Ferreira, C.R., Vianey-Saban, C., van Karnebeek, C.D.M. (eds) Physician's Guide to the Diagnosis, Treatment, and Follow-Up of Inherited Metabolic Diseases. Springer, Cham. [https://doi.org/10.1007/978-3-030-67727-5_73 DOI:10.1007/978-3-030-67727-5_73]. |
For the current overview of which pathways are covered and their status, see [https://www.wikipathways.org/index.php?title=Special:CurationTags&showPathwaysFor=Curation%3AIEM here]. | For the current overview of which pathways are covered and their status, see [https://www.wikipathways.org/index.php?title=Special:CurationTags&showPathwaysFor=Curation%3AIEM here]. | ||
| Line 6: | Line 6: | ||
To download all pathways tagged for Inborn Errors of Metabolsim as GPML, click [http://www.wikipathways.org//wpi/batchDownload.php?species=Homo%20sapiens&fileType=gpml&tag=Curation:IEM here]. | To download all pathways tagged for Inborn Errors of Metabolsim as GPML, click [http://www.wikipathways.org//wpi/batchDownload.php?species=Homo%20sapiens&fileType=gpml&tag=Curation:IEM here]. | ||
| - | For example scripts (in R), please visit [https://bigcat-um.github.io/ | + | For example scripts (in R), please visit [https://bigcat-um.github.io/IEMPathwayAnalysis/ IEMPathwayAnalysis]. |
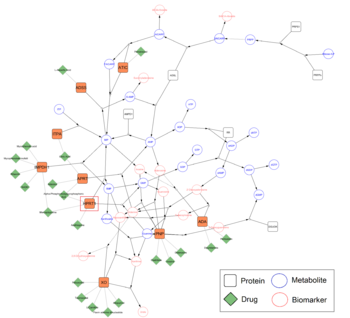
The Figure on the left shows an example of network analysis with [https://cytoscape.org/ Cytoscape], where known drug-target interactions from [https://www.drugbank.ca/ DrugBank] using the [https://cytargetlinker.github.io/ CyTargetLinker app] have been added to the [https://www.wikipathways.org/index.php/Pathway:WP4224 purine metabolism pathway]. | The Figure on the left shows an example of network analysis with [https://cytoscape.org/ Cytoscape], where known drug-target interactions from [https://www.drugbank.ca/ DrugBank] using the [https://cytargetlinker.github.io/ CyTargetLinker app] have been added to the [https://www.wikipathways.org/index.php/Pathway:WP4224 purine metabolism pathway]. | ||
Current revision
With the pathways from the Blau book (as well as with other pathways on WikiPathways), various data analysis approaches can be performed. You can find the relevant book chapter as Open Access regarding these data approaches here: Slenter, D.N., Kutmon, M., Willighagen, E.L. (2022). WikiPathways: Integrating Pathway Knowledge with Clinical Data. In: Blau, N., Dionisi Vici, C., Ferreira, C.R., Vianey-Saban, C., van Karnebeek, C.D.M. (eds) Physician's Guide to the Diagnosis, Treatment, and Follow-Up of Inherited Metabolic Diseases. Springer, Cham. DOI:10.1007/978-3-030-67727-5_73.
For the current overview of which pathways are covered and their status, see here.
To download all pathways tagged for Inborn Errors of Metabolsim as GPML, click here.
For example scripts (in R), please visit IEMPathwayAnalysis.
The Figure on the left shows an example of network analysis with Cytoscape, where known drug-target interactions from DrugBank using the CyTargetLinker app have been added to the purine metabolism pathway.