Help:Guidelines
From WikiPathways
(added refs to other guidelines) |
(refactored guidelines to simplify and more directly address pathway boundaries) |
||
| Line 4: | Line 4: | ||
= General guidelines = | = General guidelines = | ||
| - | == | + | |
| - | ==== Pathway annotation | + | === Pathway naming === |
| + | Pathways should have a descriptive name, like “Cholesterol Biosynthesis”. Do not name your pathway after your favorite pet or something else that is not descriptive of the process represented in the pathway. Do not include your name or username in the pathway name. If your pathway is specific to a particular state or tissue, this may be included in the name. If the pathway is based on content from an external source, the name of the source may be included in the name. | ||
| + | === Adding author information === | ||
| + | When creating a new pathway, add your name and contact email to the pathway. This will ensure that your information will be linked to the pathway even if it is downloaded and used in external applications. | ||
| + | === Pathway annotation === | ||
Properly annotating pathways is essential to maintaining a high quality of curation and content at WikiPathways. Annotation includes assigning the pathway to one or more categories, defining each node with a database ID and adding a description for each pathway. | Properly annotating pathways is essential to maintaining a high quality of curation and content at WikiPathways. Annotation includes assigning the pathway to one or more categories, defining each node with a database ID and adding a description for each pathway. | ||
| - | + | === References to literature === | |
Citing peer-reviewed publications is a quality standard in the scientific community and WikiPathways is no exception. We strongly encourage including relevant literature references both at the pathway level (Bibliography) and for individual interactions in pathways. | Citing peer-reviewed publications is a quality standard in the scientific community and WikiPathways is no exception. We strongly encourage including relevant literature references both at the pathway level (Bibliography) and for individual interactions in pathways. | ||
| - | + | === References to other sources === | |
For all evidence from sources other than the scientific literature, please cite the source in the Bibliography section. | For all evidence from sources other than the scientific literature, please cite the source in the Bibliography section. | ||
| - | + | === Making major edits === | |
When making major edits (deletions in particular) to an existing pathway not created by you, first suggest the changes on the pathway discussion page, as a courtesy to the original author. Starting a discussion about the proposed changes is a productive way of community editing. | When making major edits (deletions in particular) to an existing pathway not created by you, first suggest the changes on the pathway discussion page, as a courtesy to the original author. Starting a discussion about the proposed changes is a productive way of community editing. | ||
| - | + | === Content forking === | |
If you plan to add tissue- or state-specific information to a pathway describing a general process, please create a new pathway and copy the relevant content from the original pathway, rather than making edits directly to the original pathway. When naming the new related pathway, it is appropriate to use the name of the original pathway, with a suffix or addition explaining the edits. This will ensure that pathway names are not duplicated and will help in organizing related pathway content. | If you plan to add tissue- or state-specific information to a pathway describing a general process, please create a new pathway and copy the relevant content from the original pathway, rather than making edits directly to the original pathway. When naming the new related pathway, it is appropriate to use the name of the original pathway, with a suffix or addition explaining the edits. This will ensure that pathway names are not duplicated and will help in organizing related pathway content. | ||
| - | + | === Pathway boundaries === | |
| - | + | Multiple pathways may overlap or correspond to the same topic, but each should serve a unique goal in what it represents. First browse the current contents at WikiPathways to avoid duplicate work. Browsing related pathways can also give you an idea of appropriate pathway boundaries. | |
| - | + | === Other curator guidelines === | |
| - | + | Guidelines developed by other resources can also be consulted. Peter Karp and the BioCyc group, for example, provide an excellent and thorough specification to help guide all aspects of the curation of metabolic pathways: [http://bioinformatics.ai.sri.com/ptools/curatorsguide.pdf BioCyc guidelines]. We provide a [[PathwayBoundaries|paraphrased version of their rules]] defining the boundaries of metabolic pathways to serve as a sample. | |
| - | + | ||
| - | === | + | |
| - | + | ||
| - | + | ||
| - | + | ||
= Graphical style guidelines = | = Graphical style guidelines = | ||
WikiPathways does not enforce a particular graphical style for pathways. It is up to the individual author of a pathway to create a style that bests displays the relevant information. A few suggestions on specific aspects of graphical style are offered below. In addition, several other pathway resources have published guidelines on graphical style, which can serve as an inspiration for editing pathways at WikiPathways. For examples, see the [http://www.molbiolcell.org/cgi/content/full/17/1/1 MIMs], [http://www.nature.com/nbt/journal/v23/n8/abs/nbt1111.html;jsessionid=0637CAFC37BDC24F6AABD2D5A608F9E4 Kitano], and [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6TCX-416BK74-2&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=4351332fca5560eb142899161ad34b63 Pirson] guidelines. | WikiPathways does not enforce a particular graphical style for pathways. It is up to the individual author of a pathway to create a style that bests displays the relevant information. A few suggestions on specific aspects of graphical style are offered below. In addition, several other pathway resources have published guidelines on graphical style, which can serve as an inspiration for editing pathways at WikiPathways. For examples, see the [http://www.molbiolcell.org/cgi/content/full/17/1/1 MIMs], [http://www.nature.com/nbt/journal/v23/n8/abs/nbt1111.html;jsessionid=0637CAFC37BDC24F6AABD2D5A608F9E4 Kitano], and [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6TCX-416BK74-2&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=4351332fca5560eb142899161ad34b63 Pirson] guidelines. | ||
<p>In the future, WikiPathways will perform automated checking for inconsistencies in pathways, such as undefined edges, undefined nodes, non-node edge termini and inconsistent node labels etc.</p> | <p>In the future, WikiPathways will perform automated checking for inconsistencies in pathways, such as undefined edges, undefined nodes, non-node edge termini and inconsistent node labels etc.</p> | ||
| - | + | === Representing complexes === | |
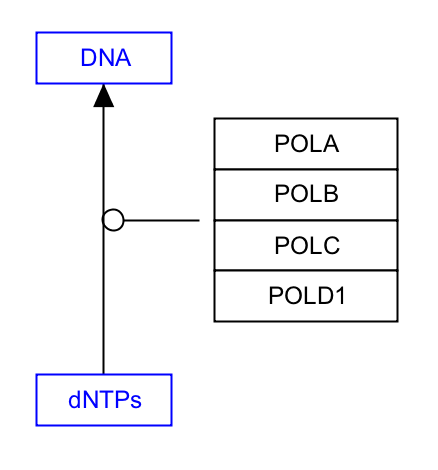
Representing molecular complexes can be done in a variety of styles. We recommend using a stacked representation whenever possible, which can easily be accomplished with the “Stack genes” feature in the editor Toolbar. The example below displays a stacked representation of the complex of subunits A, B, G and D1 of the human DNA polymerase. | Representing molecular complexes can be done in a variety of styles. We recommend using a stacked representation whenever possible, which can easily be accomplished with the “Stack genes” feature in the editor Toolbar. The example below displays a stacked representation of the complex of subunits A, B, G and D1 of the human DNA polymerase. | ||
[[image:Complex.png|left|]]<br clear="all" /> | [[image:Complex.png|left|]]<br clear="all" /> | ||
| - | + | ||
| - | + | ||
| - | + | ||
= Enforceable Policies = | = Enforceable Policies = | ||
* See [[:Help:Policies|policies page]] for list of rules regarding conduct, content and contributions at WikiPathways. | * See [[:Help:Policies|policies page]] for list of rules regarding conduct, content and contributions at WikiPathways. | ||
Revision as of 04:30, 22 May 2008
When creating or editing pathway information at WikiPathways, please keep the following guidelines in mind. These guidelines aim to promote consistency, accuracy and stability for the pathway content. By following these guidelines, you will help to maintain the highest possible quality of pathway information and to promote the success of community-based curation.
Contents[hide] |
General guidelines
Pathway naming
Pathways should have a descriptive name, like “Cholesterol Biosynthesis”. Do not name your pathway after your favorite pet or something else that is not descriptive of the process represented in the pathway. Do not include your name or username in the pathway name. If your pathway is specific to a particular state or tissue, this may be included in the name. If the pathway is based on content from an external source, the name of the source may be included in the name.
Adding author information
When creating a new pathway, add your name and contact email to the pathway. This will ensure that your information will be linked to the pathway even if it is downloaded and used in external applications.
Pathway annotation
Properly annotating pathways is essential to maintaining a high quality of curation and content at WikiPathways. Annotation includes assigning the pathway to one or more categories, defining each node with a database ID and adding a description for each pathway.
References to literature
Citing peer-reviewed publications is a quality standard in the scientific community and WikiPathways is no exception. We strongly encourage including relevant literature references both at the pathway level (Bibliography) and for individual interactions in pathways.
References to other sources
For all evidence from sources other than the scientific literature, please cite the source in the Bibliography section.
Making major edits
When making major edits (deletions in particular) to an existing pathway not created by you, first suggest the changes on the pathway discussion page, as a courtesy to the original author. Starting a discussion about the proposed changes is a productive way of community editing.
Content forking
If you plan to add tissue- or state-specific information to a pathway describing a general process, please create a new pathway and copy the relevant content from the original pathway, rather than making edits directly to the original pathway. When naming the new related pathway, it is appropriate to use the name of the original pathway, with a suffix or addition explaining the edits. This will ensure that pathway names are not duplicated and will help in organizing related pathway content.
Pathway boundaries
Multiple pathways may overlap or correspond to the same topic, but each should serve a unique goal in what it represents. First browse the current contents at WikiPathways to avoid duplicate work. Browsing related pathways can also give you an idea of appropriate pathway boundaries.
Other curator guidelines
Guidelines developed by other resources can also be consulted. Peter Karp and the BioCyc group, for example, provide an excellent and thorough specification to help guide all aspects of the curation of metabolic pathways: BioCyc guidelines. We provide a paraphrased version of their rules defining the boundaries of metabolic pathways to serve as a sample.
Graphical style guidelines
WikiPathways does not enforce a particular graphical style for pathways. It is up to the individual author of a pathway to create a style that bests displays the relevant information. A few suggestions on specific aspects of graphical style are offered below. In addition, several other pathway resources have published guidelines on graphical style, which can serve as an inspiration for editing pathways at WikiPathways. For examples, see the MIMs, Kitano, and Pirson guidelines.
In the future, WikiPathways will perform automated checking for inconsistencies in pathways, such as undefined edges, undefined nodes, non-node edge termini and inconsistent node labels etc.
Representing complexes
Representing molecular complexes can be done in a variety of styles. We recommend using a stacked representation whenever possible, which can easily be accomplished with the “Stack genes” feature in the editor Toolbar. The example below displays a stacked representation of the complex of subunits A, B, G and D1 of the human DNA polymerase.
Enforceable Policies
- See policies page for list of rules regarding conduct, content and contributions at WikiPathways.