Help:Guidelines
From WikiPathways
For the latest curation guidelines, see our Ten Simple Rules paper.
General guidelines
Pathway content
- All contents of a pathway, including entities of the graphical representation (data node labels, labels, comments) and description, should be in English.
Pathway naming
- Pathway names should be in English.
- Use sentence case. Write words as they appear in a sentence, for example "Activation of NLRP3 inflammasome by SARS-CoV-2", using upper case for the first word only.
- Use "signaling" instead of "signalling".
- Pathways should have a descriptive name, like “Cholesterol biosynthesis”.
- Do not include your name, username, species names or species codes in the pathway name.
- Try to pick a name that is unique at WikiPathways.
- If your pathway is specific to a particular state or tissue, this may be indicated in the name.
- If the pathway is based on content from an external source, the name of the source may be indicated in the name.
Pathway annotation
Properly annotating pathways is essential to maintaining a high quality of curation and content at WikiPathways. Annotation includes adding one or more ontology tags, defining each node with a database ID and adding a description for each pathway.
Content forking
If you plan to add tissue- or state-specific information to a pathway describing a general process, please create a new pathway and copy the relevant content from the original pathway, rather than making edits directly to the original pathway. When naming the new related pathway, it is appropriate to use the name of the original pathway, with a suffix or addition explaining the edits. This will ensure that pathway names are not duplicated and will help in organizing related pathway content.
Pathway boundaries
Multiple pathways may overlap or correspond to the same topic, but each should serve a unique goal in what it represents. First browse the current contents at WikiPathways to avoid duplicate work. Browsing related pathways can also give you an idea of appropriate pathway boundaries.
Author information and referencing pathway sources
Author information on pathways
When creating a new pathway, your name as designated in your profile will automatically be added to the Pathway Page. To ensure that your information is linked to the pathway even if it is downloaded and used in external applications, you should also add your name and contact information to the Information Area of the pathway. This is done in the editor by selecting the Information Area and editing the relevant information in the Properties panel.
References to literature
Citing peer-reviewed publications is a quality standard in the scientific community and WikiPathways is no exception. We strongly encourage including relevant literature references both at the pathway level and for individual interactions in pathways.
References to other sources
For all pathway content from sources other than the scientific literature (for example commercial software products), please cite the source in the Bibliography section and add an description of the original source in the Description section. It is also a good idea to check the EULA (End User License Agreement) for any external source of pathway content.
Curator etiquette
Making major edits
When making major edits (deletions in particular) to an existing pathway not created by you, first suggest the changes on the pathway Discussion page, as a courtesy to the original author. Starting a discussion about the proposed changes is a productive way of community editing.
Pathway curation guidelines from external resources
Guidelines developed by other resources can also be consulted. While much of the material may be specific to their curation tools, there are general rules that apply to any pathway curation effort... where sloppy biology must be systematically modeled and ultimately displayed on a page.
- Peter Karp and the BioCyc group provide an excellent and thorough specification to help guide all aspects of the curation of metabolic pathways: BioCyc guidelines.
- Reactome hosts a wiki where they've posted the Reactome Curator Guide
- Kitano H, Funahashi A, Matsuoka Y, Oda K: Using process diagrams for the graphical representation of biological networks. Nat Biotechnol 2005, 23(8):961-966. pubmed
- Pirson I, Fortemaison N, Jacobs C, Dremier S, Dumont JE, Maenhaut C: The visual display of regulatory information and networks. Trends Cell Biol 2000, 10(10):404-408. pubmed
- Kohn KW, Aladjem MI, Weinstein JN, Pommier Y: Molecular interaction maps of bioregulatory networks: a general rubric for systems biology. Mol Biol Cell 2006, 17(1):1-13. pubmed
Curator guidelines
Below are our recommendations on specific aspects of editing that best utilize the tools and data model. In terms of graphical style, it is up to the individual author of a pathway to create a style that best displays the relevant information. WikiPathways does not enforce a particular graphical style.
Recommended use of pathway elements
For the recommended use of the most commonly used elements of the WikiPathways editor palette, see our Editor Palette Guide.
Illustrating molecular states
WikiPathways allows for the addition of a state to any data node, to illustrate post-translational modifications (PTMs), mutations etc. To add a state to a data node, right-click on the node and select Add State...:
The state label is customizable, as is its color, size and placement. Data nodes can have more than one state.
Post-translational modifications
Although data cannot yet be mapped to states in WikiPathways/PathVisio, it is possible in Cytoscape. To facilitate data mapping, we recommend adding a descriptive comment to the state that can be used to setup a visual style in Cytoscape. This comment should include an identifier for the parent protein, the amino acid position, amino acid, type of post translational modification and up- or down-regulation. For example, to map phosphoprotein abundance data, the following comment indicates an upregulation phosphorylation event on threonine 491 in protein P04049:
parent=P04049; parentsymbol=RAF1; site=IGDFGLAtVksRWsG; position=thr491; sitegrpid=447748; ptm=p; direction=u
In this particular case, the comment includes the PhosphoSite Plus site group id, the flanking sequence and the parent symbol as well. To add a comment, double-click on the state and enter text in the Comments tab.
Mutation status
In some pathways, it may be useful to indicate a genes mutation state in the particular conditions or disease described, for example in pathways summarizing cancer processes. We recommend using the following convention for using states to illustrate mutation state:
An example of a pathway using states to illustrate mutation status can be found here.
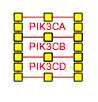
Representing paralogs and complexes
Representing groups of paralogs and complexes can be done in a variety of styles. We recommend using a stacked representation whenever possible, which can easily be accomplished with the “Stack genes” feature in the editor Toolbar. In the example below, the human RNA Polymerase I subunits are represented as a stack.
You will also likely want to "group" the stacked genes. Once selected, genes can be grouped by pressing CTRL-G, or choosing 'group' in the right-click menu. In the case of complexes, however, you will want to specifically choose "Create Complex" (CTRL-P) to create a visual and properly modeled representation of a molecular complex.
| Before | After |
|---|---|
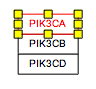
| 
|
Representing reactions
- When two reaction steps follow each other in a pathway (i.e. the product of reaction A is the substrate for reaction B), it is best to use a single element for this entity.
- On the other hand, common entities that are involved in many reactions may be drawn as multiple entities for clarity, so you don't get a large messy hairball. Common metabolites include H2O, H+, ATP, NADH.
- Certain common metabolites that are unimportant for understanding of the pathway as a whole, such as H2O and H+ may even be left out altogether.
Representing enzymatic reactions
In order to properly store enzymatic reactions in the WikiPathways data model, place a line anchor on the line connecting the two reactants, then use a second connector from the enzyme to the anchor to denote the enzymatic interaction. To add an anchor to a line, right-click on the line and select 'Add Anchor'. Ideally, use for the line connecting the enzyme and the anchor the MIM-interaction type "Catalysis", giving the circle near the anchor point.
| Enzymatic reaction |
|---|
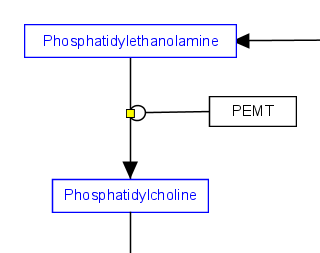
|
Stoichiometry
The stoichiometry of reactions is not usually modeled in wikipathways. If you wish you can add this information as a comment attached to a reaction or you can use the label for a metabolite to indicate stoichiometry, as suggested below.
| Stoichiometry in metabolite labels |
|---|
|
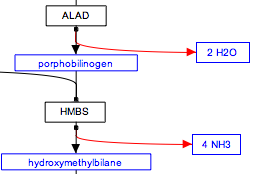
Representing multiple similar reactions
Representing multiple similar reactions so that they are properly stored in the underlying data model can sometimes present a challenge. Below is an example of using a connector hub (a line anchor) to display this. Right-click on any line to add a line anchor, then connect multiple lines for different reactions to this hub.
| Wrong | Right! |
|---|---|
| 
|
Using labels
To enter text for a label, double-click on the label (or choose 'Properties' in the right-click menu) and use the Label Text area to type the label, including line returns. The label can consist of multiple lines of text. You can also outline a label by choosing 'Rectangle' or 'RoundedRectangle' from the 'Outline' field in the properties panel
| Outlined label |
|---|
|
Representing metabolites
Metabolites and small molecules should not be represented as a label, but rather as a data object connected to a database entry (like genes and proteins). Use the 'Met' button in the editor to create a metabolite object. Double-click the object to fill in the text label, identifier and database, or simply search by name or identifier. While at it, consider using an anchor and Catalysis interaction too.
| Wrong | Right! | Even better! |
|---|---|---|
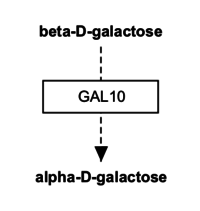
| 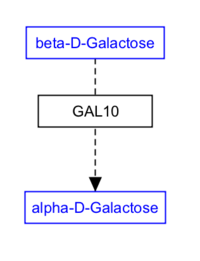
| 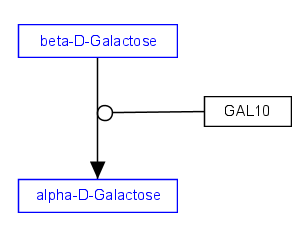
|
Representing interactions
Complicated connections involving curves or corners can easily be managed using "smart connectors". Any line or arrow can be made to curve or bend by choosing 'Curved','Elbow' or 'Segmented' for 'Line type' in the right-click menu. Connect the ends of the line to their targets and the connector will bend where necessary. You can select the line again to adjust the way points of the path (blue diamond handles).
| Using elbows |
|---|
|
Replace arc/arrowhead combinations with curved or elbowed connectors.
| Using curves |
|---|
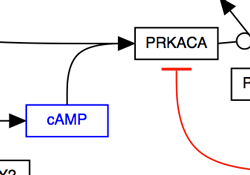
|
These options should help you style your pathway so that it is easy to "read" and has a layout that is familiar to any biologist or biochemist.
To make sure you've connected all your lines, press CTRL-L to reveal all unlinked line ends and highlight them in green.
| Before | After |
|---|---|
| 
|
Representing cellular compartments
Representing cellular compartments as graphics is a useful way to illustrate location spatial relationships between objects and where processes take place. While cellular compartments can be illustrated in a number of ways, we recommend using an oval or line (or double line) with line thickness of 5 and colored light grey. In the example below, the nucleus is depicted in this way.
Example pathways
The content at WikiPathways is diverse in both style and content. In addition to the above graphical style guidelines, we have identified a list of example pathways that are considered well-formed in terms of annotation, representing interactions and overall layout.
- Apoptosis (Homo sapiens)
- One carbon metabolism (Homo sapiens)
- Long-Day Flowering Time Pathway (Arabidopsis thaliana)
- Glycolysis and gluconeogenesis (Homo sapiens)
You can find a list of all Featured pathways here.
Inclusion of pathways into specific pathway collections
WikiPathways uses quality tags to identify subsets of pathways as specific collections. For example, the quality tags "Featured version" and "Approved version" identify subsets of pathways in this way. Using tags to identify pathways as belonging to a specific collection is informative when viewing pathways, and also facilitates downloading of pathway archives. For inclusion to the archives mentioned above the criteria are as follows:
- Approved: The "Approved version" contains pathways that are considered to be complete, or close to complete, in terms of the graph representation. This means that **all interactions must be connected to pathway entities**, and **data nodes must be annotated** when possible. While "Approved version" pathways may not be complete in terms of bibliography and description, they are still useful for data analysis in other programs, like PathVisio and Cytoscape.
- Featured (discontinued): The "Featured version" collection contains pathways that represent the highest quality in terms of annotation. A pathway tagged as "featured" has a description and is annotated with ontology tags. The pathway also has to have a proper graph representation, meaning all relevant interactions must be connected to pathway entities and all data nodes must be annotated, at least for gene products. NOTE: This collection is no longer actively being updated/maintained.
NOTE: Pathways that don't depict a biological process, for example list-based pathways, should NOT be included in the Featured and Approved sets.
Enforceable Policies
See policies page for list of rules regarding conduct, content and contributions at WikiPathways.