Portal:Lipids
From WikiPathways
(Disabled the logo box for now and moved News to balance.) |
Current revision (19:02, 9 September 2020) (view source) |
||
| (2 intermediate revisions not shown.) | |||
| Line 10: | Line 10: | ||
<div style="float:left; width:62%"> <!-- This width add to the the margin below to equal 99%--> | <div style="float:left; width:62%"> <!-- This width add to the the margin below to equal 99%--> | ||
| + | |||
| + | {{{{FULLPAGENAME}}/box-normal | ||
| + | |<!-- TITLE OF BOX --> | ||
| + | Data access | ||
| + | |<!-- EDIT LINK --> | ||
| + | {{FULLPAGENAME}}/DownloadPWs | ||
| + | |<!-- CONTENT LINK --> | ||
| + | {{FULLPAGENAME}}/DownloadPWs | ||
| + | |<!-- FOOTER LINK --> | ||
| + | }} | ||
{{{{FULLPAGENAME}}/box-normal | {{{{FULLPAGENAME}}/box-normal | ||
| Line 59: | Line 69: | ||
}} | }} | ||
--> | --> | ||
| + | |||
| + | [[Category:Portals]] | ||
Current revision
Welcome to the Lipids Pathways Portal on WikiPathways
This portal is set up to coordinate the development of lipid pathways and get together the community interested in interactive lipid pathways.
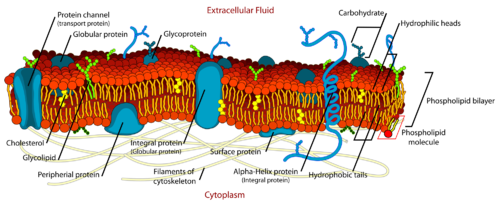
Lipids serve several important biological purposes, such as providing membrane structure, and signalling.
Thanks to the LIPID MAPS team for sharing their pathway knowledge through WikiPathways!
For more details on the LipidMaps pathways, please read the following paper: "A Mouse Macrophage Lipidome" Edward A. Dennis et.al. J Biol Chem. 2010 Dec 17; 285(51): 39976-39985.[1]
Read more about the Scalable Curation grant we received from Elixir together with the LIPID MAPS team, or on the EpiLipidNet Cost Action.
This portal is also featured in the 2021 NAR Database Issue on WikiPathways and Metabolomics 2021 Review Article on Interpreting the lipidome.
Data access
| RDF | API |
|---|---|
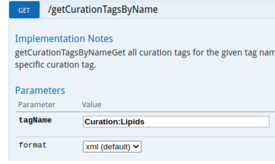
| Data can be obtained using SPARQL queries. An example to obtain all Proteins from the Lipids Community: | The WikiPathways Webservice (API) allows for automated retrieval of Pathway content (in several formats, e.g. json, XML, html, GPML). |
 | 
|
| Example SPARQL Query | API Call Lipids Community |
Pathways
Existing pathways
Pathways which have been approved for data analysis can be found on the Featured pathway page. We have split these up in 7 groups (Sphingolipids, Eicosanoids, Glycerolipids and Glycerophospholipids, Phosphatidylglycerides, Fatty Acids, Cholesterol, General Pathways related to Lipids).
Extend existing (approved) pathways with new literature
- More proteins, localisation, details on regulation of (endocytic) membrane traffic [2] and [3] by phosphoinositide 3-kinases, and activity and tumor suppression thereof for WP4971.
Update existing (not yet approved) pathways
| Sphingolipid Metabolism (integrated pathway) (mouse) |
| Elongation of (very) long chain fatty acids (mouse) |
Create new pathways
- Public resources for pathway information: LIPID MAPS and SwissLipids.
- See our Help section for New Contributor QuickStart, Editor's Palette Guide and Guidelines for Contributors.
Start a new pathway!
- Phosphoinositide Signaling
- PI3-K in Human diseases
- Class I PI3-K Isoforms in Endometrial Cancer
- Inositol trisphosphate receptor
Contact us
Send questions about WikiPathways to the wikipathways-discuss mailing list.
How to add a pathway to the portal
The list of Featured Pathways is not static and can be updated at any time. If you know of a pathway that should be added, please contact the administrator.
Featured Pathway
|
Metabolism of sphingolipids in ER and Golgi apparatus (Homo sapiens) Image does not exist Metabolism of sphingolipids in ER and Golgi apparatus |
| View all Featured Pathways for this Portal |
News
2023-03-16 - Fully curated version of the Bacterial ceramide synthesis pathway WP5271 has been released!
2022-12-22 - Find the curation reports [here](https://egonw.github.io/lipidmaps-wp-curation/), and help update our lipid pathways!
2022-09-21 - Fully curated version of the Ether lipid synthesis pathway WP5275 has been released! (data supplied by Robert Murphy)
2022-07-16 - Seven new lipid signaling pathways, and three new metabolic lipid pathways have been added to this portal!
2021-06-23 - A new pathway on 7-oxo-C and 7-beta-HC pathways (WP5064) with data from Fig.4 and 5 from Griffiths et al (2020) has been created.
2021-02-26 - We have extended the pathway on Oxysterols derived from cholesterol (WP4545) with data from Fig.1, 2 and 3 from Griffiths et al (2020).
2020-10-13 - The EpiLipidNET project has started. See Twitter and the COST action info page.
2020-10-01 - A brand new pathway on Phosphoinositides metabolism (WP4971) is now available (Human).
2019-10-23 - All Mouse pathways (originally from LipidMaps) have been converted to Human! See WP4718 - WP4726 .
2019-08-21 - Pathway:WP4350 and Pathway:WP4351 on Omega 3 & 6 FA synthesis and Omega 9 and saturated FA synthesis (mouse) have been updated!
2019-08-19 - Pathway:WP4344 and Pathway:WP4690 on Sphingolipids (general overview) and Sphingolipids (integrated pathway) have been updated!
2019-08-16 - Pathway:WP4345 on Glycerolipids and Glycerophospholipids has been updated!
2019-01-29 - Pathway:WP4348 and Pathway:WP4349 on Eicosanoid metabolism by LOX and CYP (mouse) have been updated!
2019-01-25 - Pathway:WP4347 on Eicosanoid metabolism by COX (mouse) has been updated!
2019-01-21 - Pathway:WP4346 on Cholesterol metabolism (mouse) has been updated!
2019-01-11 - More pathways were added to the featured pathway list.
2018-10-11 - This portal was created.